Researchers of Dr. Ruhong Zhou's team have developed a combined experimental and theoretical approach to discover two new optimal ephithelial cellular adhesion molecule (EpCAM) aptamers.
Description:
Given the great promise of these nucleic acid aptamers in therapeutic applications, Zhou and his team applied it to discover two new optimal epithelial cellular adhesion molecule (EpCAM) aptamers. The structure-based in silico method first predicts their binding modes and then optimizes them for EpCAM with molecular dynamics simulations, docking, and free energy calculations. The isothermal titration calorimetry (ITC) experiments further confirm that the newly designed EpCAM aptamers indeed exhibit enhanced affinity over a previously patented nanomolar aptamer, EP23. Moreover, the study suggests that EP23 and the de novo designed aptamers primarily bind to EpCAM dimers (and not monomers, as hypothesized in previous published works), suggesting a new paradigm for developing EpCAM-targeted therapies.
The content below presents a part of their research findings:
EP23 Structure and Mg2+
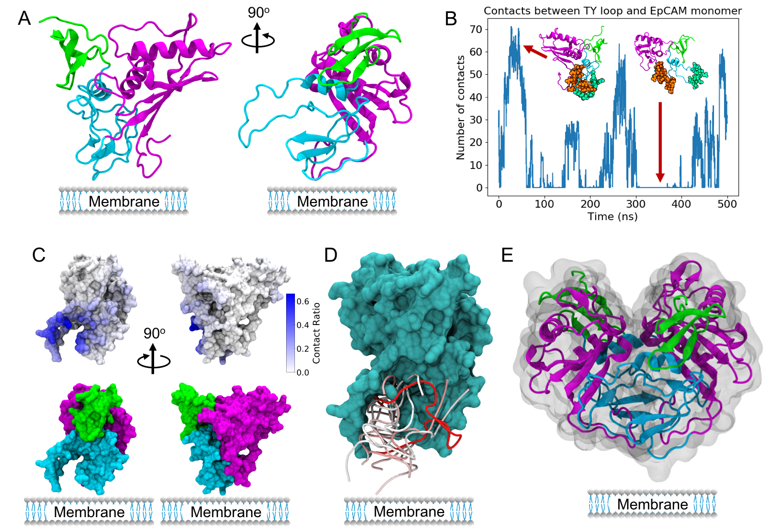 Figure 1
Figure 1
(A) The Mfold-predicted secondary structure of EP23. (B) The 3-D folded structure of EP23 determined from MD simulations with the 4 base pairs predicted from Mfold highlighted in blue. (C) Base pair dynamics of the Mfold predicted secondary structure. The terminal base pair (blue) changed conformations slightly from a Watson-Crick to a Hoogsteen-like configuration. (D) Mg2+ ions binding nucleotides 8 (top) and 18 (bottom) over the course of the MD simulations. Two long-lived bound ions are circled in red. (E) Mg2+ ion contact ratio mapped onto the EP23 aptamer surface. The two bound Mg2+ ions correspond to the long-lived bound ions in (D).
EP23-EpCAM monomer binding results:
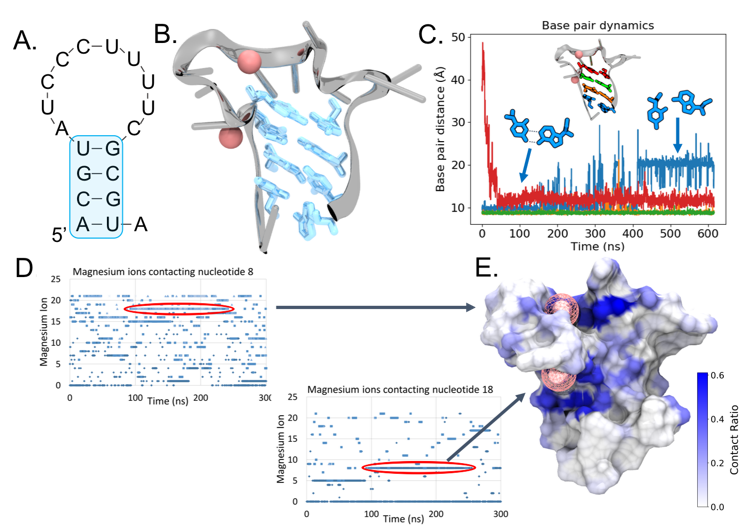
Figure 2
(A) EpCAM monomer showing the N-terminal domain in green, TY loop in blue and C-terminal domain in magenta. (B) EpCAM TY loop fluctuations observed in control EpCAM explicit solvent MD simulations. The residues used for contact analysis are shown as spheres in green (TY loop) and brown (core EpCAM). (C) EpCAM mapped contacts from the top 200 Dot2.0 docking conformations, showing the strong preference for TY loop binding conformations. (D) Top 10 docking conformations predicted by Dot2.0, all targeting the TY loop region at the dimer interface and the membrane side of the protein. The RNA backbone is shown, colored by energy from strong (red) to weak (blue). (E) EpCAM dimer crystal structure, showing dimerization using the TY loop.
In this paper, Zhou's team also depicted the results of EP23 binding EpCAM dimer and EP23 mutation binding affinity. More details can be found from the orginal work.
In a conclusion, with the current limitations of the numerous RNA structure prediction models, this physics -based RNA structure-building framwork allowed for appropraite fluctuations in the RNA structure while still identifying stable, robust RNA conformations. The research results were largely based on Molecular Dynamics Simulations, Free Energy Perturbation Calculations, and DOT2 Docking.
The research paper was first published in PNAS (Preceedings of the National Academy of Sciences of the United States of America), March 31st, 2020.
Please view the entire paper by referring to the orginal link:
https://www.pnas.org/content/117/15/8486
About SIAS
Shanghai Institute for Advanced Study of Zhejiang University (SIAS) is a jointly launched new institution of research and development by Shanghai Municipal Government and Zhejiang University in June, 2020. The platform represents an intersection of technology and economic development, serving as a market leading trail blazer to cultivate a novel community for innovation amongst enterprises.
SIAS is seeking top talents working on the frontiers of computational sciences who can envision and actualize a research program that will bring out new solutions to areas include, but not limited to, Artificial Intelligence, Computational Biology, Computational Engineering and Fintech.

