Polyamines are a class of compounds containing two or more amino groups, and in human cells, the precise regulation of polyamine homeostasis is essential to maintening cellular health. Polyamine homeostasis depends on two systems: the metabolic system, which regulates the production and degradation of polyamines, and the transport system, which determines the distribution of polyamines in the cell. The lysosomal transporter ATP13A2 exports polyamines from the lysosome to the cytoplasm and plays a critical role in balancing the concentration of polyamines between the cytoplasm and the lysosome. It had been discovered that abnormalities in the human ATP13A2 (hATP13A2) gene was associated with a number of diseases, including Kufor-Rakeb Syndrome (a rare autosomal recessive form of juvenile-onset atypical Parkinson disease) and Autosomal Recessive Spastic Paraplegia Type 78. Although ATP13A2 plays a key role in polyamine homeostasis, its molecular mechanism is still unclear. Recently, Professor Ruhong Zhou from College of Life Sciences, Zhejiang University and Shanghai Institute of Advanced Study, Zhejiang University and collaborators jointly published a study titled Conformational cycle of human polyamine transporter ATP13A2 in Nature Communications. The authors captured high-resolution atomic structures of almost the entire conformational cycle of the hATP13A2 polyamine transporter by cryo-electron microscopy (Figure 1), and performed molecular dynamics simulations and mass spectrometry to reveal the molecular mechanism of polyamine transport to reveal structural basis for the recruitment, transport, and release of human ATP13A2 polyamine substrates. The work expanded out understanding in pathogenic mechanism of hATP13A2 in neuronal diseases and would facilitate drug design.
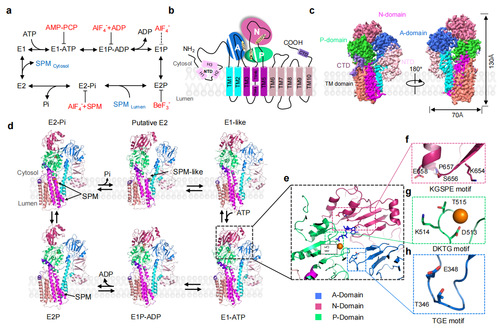
Figure 1. Cryo-EM analysis of human ATP13A2
1. Polyamine recruitment. The first step in transporting substrates from the luminal side to the cytoplasmic side is the recruitment of polyamines to the hATP13A2 protein. The team identified a narrow binding pocket on the luminal side of the endo/lysosome with an outward-opening structure in the cryo-electron microscopy E2P structure of hATP13A2. Molecular dynamics simulations demonstrated that this is the entry site for polyamine recruitment.
2. Polyamine transport. With the energy release of dephosphorylation hydrolysis, phosphorylated hATP13A2 switches from the E2P state to the E2-Pi state, driving the movement of substrates from the cytosolic side to the other side. Although the cryo-electron microscopy structure did not capture the exact binding site of the polyamine here, the team confirmed the second substrate binding site of the polyamine is on the intracellular sidevia through coarse-grained molecular dynamics simulations, mutation function experiments, and cross-linked mass spectrometry. This is the first capture of the intermediate state in the polyamine transport.
3. Polyamine release. Accompanied by the release of phosphate ions, hATP13A2 enters the E2 state prior to substrate release. The team observed a large number of more dynamic lipid analogs around substrate release pocket in the cryo electron density map. The authors then examined the interaction of lipids and polyamines through coarse-grained molecular dynamics simulations (Figure 2) and found that these lipids were most likely strongly negatively-charged phospholipids, e.g., phosphatidylinositol 4,5-bisphosphate (PIP2). Finally, using molecular dynamics simulations that accounted for all atoms, the authors identified a third, highly hydrated polyamine-binding site, revealing the complete polyamine transport pathway.
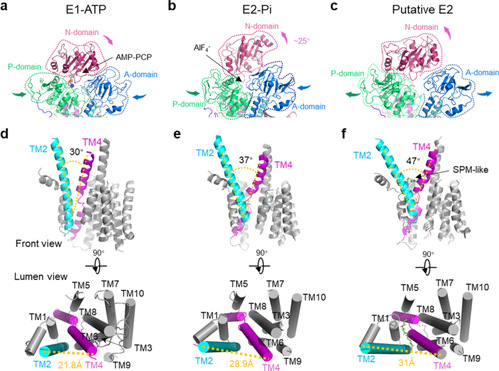
Figure 2. Conformational arrangements among intermediate states
In summary, the work resolved and modeled a series of conformational states of the full conformational cycle of the human polyamine transporter protein ATP13A2 via structural and computational biology methods and revealed the transport mechanism of the substrate polyamine molecule (Figure 3), providing guidance for subsequent structure-based drug design.
To learn more about the work please visit https://doi.org/10.1038/s41467-023-37741-0.
About Professor Ruhong Zhou
Ruhong Zhou, AAAS Fellow, APS Fellow, is currently Qiushi Chair Professor, Dean, College of Life Sciences; Dean, Shanghai Institute for Advanced Study; and Director, Institute of Quantitative Biology of Zhejiang University. He is also an Adjunct Professor at Department of Chemistry, Columbia University. Before that, he was a Distinguished Research Staff Scientist, and Head of the Soft Matter Science Department at IBM Research.
Professor Zhou’s research focus on Quantitative Biology; Machine Learning, Deep Learning in Biology; Biophysics; and Bio-Nano Interface (nanomedicine). He has authored and co-authored more than 300 journal publications with 25000+ total citations (Google H-index 81), filed 32 international patents and delivered 300+ invited talks at major conferences and universities worldwide. He is part of the IBM BlueGene team that won the 2009 National Medal on Technology (presented by President Obama). He has won the IBM Outstanding Technical Achievement Award (the highest technical award within IBM; 10 times), the IBM Outstanding Innovation Award (twice), and many IBM Research Division Awards. He also won the American Chemical Society DEC Award on Computational Chemistry. He was elected to AAAS Fellow (American Association of Advancement of Science) and APS Fellow (American Physical Society) in 2011. He received his PhD in Biophysical Chemistry from Columbia University, MS in Condensed Matter Physics and BS in Physics both from Zhejiang University.
About SIAS
Shanghai Institute for Advanced Study of Zhejiang University (SIAS) is a jointly launched new institution of research and development by Shanghai Municipal Government and Zhejiang University in June, 2020. The platform represents an intersection of technology and economic development, serving as a market leading trail blazer to cultivate a novel community for innovation amongst enterprises.
SIAS is seeking top talents working on the frontiers of computational sciences who can envision and actualize a research program that will bring out new solutions to areas include, but not limited to, Artificial Intelligence, Computational Biology, Computational Engineering and Fintech.

