The outer membrane structure is common in Gram-negative bacteria, mitochondria and chloroplasts, and contains outer membrane β-barrel proteins (OMPs) that are essential interchange portals of materials. The folding of the tertiary structure of β-barrel proteins is critical to their function: new peptide chains are synthesized intracellularly and transported to the outer membrane, where the β-barrel assembly machinery (BAM) helps fold them into a barrel-shaped structure and integrate them into the outer membrane. All known OMPs share the antiparallel β-strand topology, implicating a common evolutionary origin and conserved folding mechanism. Recent years witnessed emerging of the study on structure and function of BAM in structural biology. The BAM proteins have been found to initiate a dynamic molecular model of outer membrane protein folding through the lateral opening of the core subunit BamA's own β-barrel by using the interaction of the first β-sheet of BamA with the last β-sheet (signal peptide) of the substrate OMP without consuming energy. Models have been proposed for bacterial β-barrel assembly machinery (BAM) to initiate OMP folding, but mechanisms by which BAM proceeds to complete OMP assembly remain unclear.

On 26th April 2023, Professor Ruhong Zhou and collaborators published the article Structural basis of BAM -mediated outer membrane β-barrel protein assembly in Nature, reporting intermediate structures of BAM assembling an OMP substrate, EspP, demonstrating sequential conformational dynamics of BAM during the late stages of OMP assembly, which is further supported by molecular dynamics simulations. Mutagenic in vitro and in vivo assembly assays reveal functional residues of BamA and EspP for barrel hybridization, closure and release. The work provides novel insights into the common mechanism of OMP assembly.
It is challenging to capture the intermediate complexes of BAM-integrated substrates due to the rapid assemble of BAM substrates in vivo. The authors succeeded in obtaining BAM-EspP complex proteins by inserting cysteines at different positions in the predicted interactions between BAM and EspP to allow spontaneous formation of disulfide bonds. For the first time they resolved BAM-EspP conformations using single-particle cryo-electron microscopy. The observation reveals Cryo-EM structures of intermediate assembly complexes of BAM and the substrate EspP.
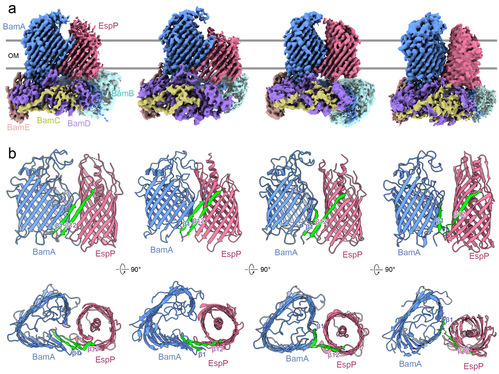
The conformational changes of the BamA barrel during state transition are accompanied by the rotation of the periplasmic ring formed by the POTRA domains of BamA and BamBCDE. Nascent substrate OMP inserts through the barrel of BamA from the C terminus, and the C-terminal β-strand of the substrate OMP then forms an anti-parallel β-seam with the N-terminal β1 of BamA at the lateral opening. The substrate OMP continues to fold into anti-parallel β-sheets from the hybrid barrel interface, and substrate OMP closes into a barrel through interactions between the N-terminal β-strand and the overhang of the C-terminal β-strand. Then BamA also closes its barrel by rolling its N-terminal strands towards its C terminus to peel off from the hybrid interface, breaking the interactions with the substrate OMP towards the periplasmic end. In the end, the hybrid barrels rotates 180 degree along the y axis showing the interface between the N terminus of substrate OMP and the C terminus of BamA, and BAM and the assembled substrate OMP are separated.
The conformational synchronization and interactive coupling between the barrel and periplasmic domains support a 'rotation-and-closing' mechanism for BAM, which agrees with and complements the rotation-and-insertion mechanism proposed for the initiating stage of OMP assembly.

The work reveals structural details of BAM-mediated barrel closure and release, which, together with other reports, provides an insightful understanding of the complete assembly mechanism of OMPs. These findings may also lead to the discovery of functional inhibitory targets for developing effective antibacterial agents.To access the work please visit https://www.nature.com/articles/s41586-023-05988-8.
About Professor Ruhong Zhou
Ruhong Zhou, AAAS Fellow, APS Fellow, is currently Qiushi Chair Professor, Dean, College of Life Sciences; Dean, Shanghai Institute for Advanced Study; and Director, Institute of Quantitative Biology of Zhejiang University. He is also an Adjunct Professor at Department of Chemistry, Columbia University. Before that, he was a Distinguished Research Staff Scientist, and Head of the Soft Matter Science Department at IBM Research.
Professor Zhou’s research focus on Quantitative Biology; Machine Learning, Deep Learning in Biology; Biophysics; and Bio-Nano Interface (nanomedicine). He has authored and co-authored more than 300 journal publications with 25000+ total citations (Google H-index 81), filed 32 international patents and delivered 300+ invited talks at major conferences and universities worldwide. He is part of the IBM BlueGene team that won the 2009 National Medal on Technology (presented by President Obama). He has won the IBM Outstanding Technical Achievement Award (the highest technical award within IBM; 10 times), the IBM Outstanding Innovation Award (twice), and many IBM Research Division Awards. He also won the American Chemical Society DEC Award on Computational Chemistry. He was elected to AAAS Fellow (American Association of Advancement of Science) and APS Fellow (American Physical Society) in 2011. He received his PhD in Biophysical Chemistry from Columbia University, MS in Condensed Matter Physics and BS in Physics both from Zhejiang University.
About SIAS
Shanghai Institute for Advanced Study of Zhejiang University (SIAS) is a jointly launched new institution of research and development by Shanghai Municipal Government and Zhejiang University in June, 2020. The platform represents an intersection of technology and economic development, serving as a market leading trail blazer to cultivate a novel community for innovation amongst enterprises.
SIAS is seeking top talents working on the frontiers of computational sciences who can envision and actualize a research program that will bring out new solutions to areas include, but not limited to, Artificial Intelligence, Computational Biology, Computational Engineering and Fintech.

