The SARS-CoV-2 global epidemic has led to over 800 million infections and nearly 7 million deaths, posing a major threat to human life and health and altering the global political, economic and public health landscape. SARS-CoV-2 is a Positive-Sense Single-Stranded RNA (+ssRNA) Virus, with the viral RNA genome itself encoding both the genetic information and regulate viral genome replication. It is anticipated that RNA viruses would co-opt host RNA-binding proteins (RBPs) to perform their particular functions during transcription and replication. For instance, several host RBPs have been identified that affect the recruitment of plus viral strands for replication, play important roles during the switch from translation to replication, control viral RNA synthesis, affect the stability of viral RNAs, and participate in multiple events during infection. Therefore, a deeper understanding of the interactions between viral RNAs and host RBPs will provide new insights into the virus life cycle and help the development of novel antiviral strategies.
Professor Ruhong Zhou from Shanghai Institute for Advanced Study, Zhejiang University published the article ‘Mutagenesis and structural studies reveal the basis for the specific binding of SARS-CoV-2 SL3 RNA element with human TIA1 protein’ in Nature Communications on 22 Jun 2023, illustrating the binding mechanism of SL3, a conservative cis-acting element in the 5'-UTR of the SARS-CoV-2 RNA genome, and human TIA1 protein via acombined theoretical and experimental approach including molecular modeling, free energy calculations, electrophoretic mobility shift assay (EMSA), and antisense oligonucleotides (ASO) assays. These findings open an avenue to explore the viral RNA-host protein interactions and provide a pioneering structural basis for RNA-targeting antiviral drug design.
First, the authors combined RNA structure prediction with gel EMSA to confirm the high affinity (Kd=0.78±0.09µM) of the SL3 element in the SARS-CoV-2 RNA genome when binding with human TIA1 protein. The three-dimensional structure of the SL3-TIA1 binding complex was constructed by molecular modelling and molecular dynamics simulations (Figure 1), which revealed that aromatic ring stacking, hydrogen bonding and hydrophobic interactions in addition to electrostatic interactions between the positively charged amino acid residues and the negatively charged RNA backbone, contribute to the specific binding of SL3 with TIA1 protein.
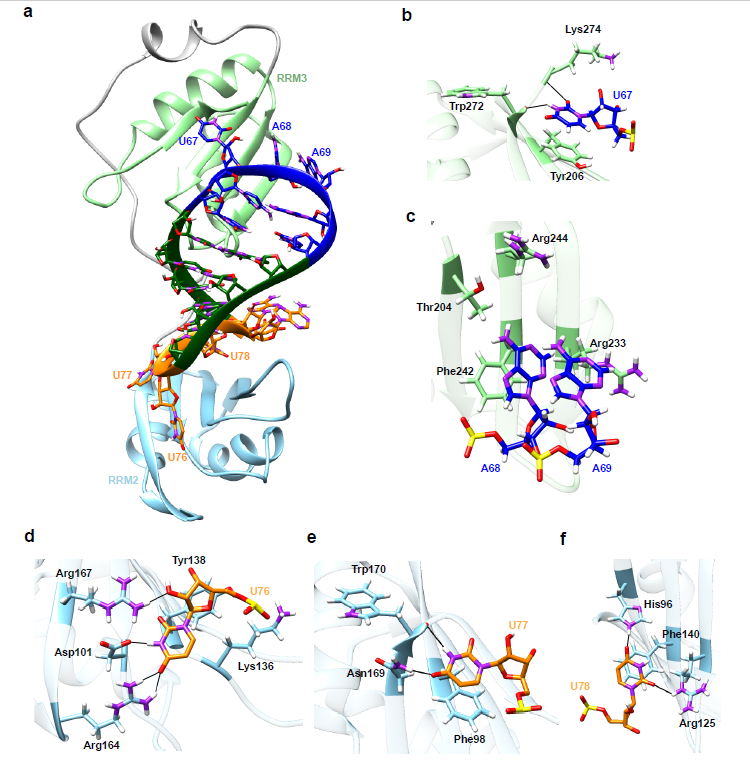
Figure 1. The putative 3D binding model for SARS-CoV-2 SL3 RNA element and human TIA1 complex. (a) Overview of the most probable 3D structure of binding complex constructed by computational modeling. TIA1 RRM2, RRM3, and flexible linker are colored by sky blue, light green, and gray, respectively. Hairpin loop, stem, and terminal loop of SL3 are separately colored by blue, green, and orange. Details of interactions between TIA1 RRM3 and nucleotides (b) U67, (c) A68 and A69 in hairpin loop region of SL3 element. Details of interactions between TIA1 RRM2 and nucleotides (d) U76, (e) U77, and (f) U78 in 3′-terminal loop of SL3. Atoms of phosphorus, oxygen, nitrogen, and hydrogen are colored yellow, red, purple, and white, respectively, and carbon atoms are colored according to their locations. Hydrogen bonds between RNA nucleotides and TIA1 residues are indicated by thin black lines. Rotations may be adopted to display the interaction details in subfigures (b–f).
Furthermore, by designing various SL3 variants with truncated loop lengths or sequence alterations and combining them with subsequent EMSA experiments, the authors found that the 7-nt hairpin loop and the 5-nt 3'-terminal loop in SL3 are essential for binding. As shown in Figure 2, truncation or sequence alteration of the hairpin and 3'-terminal loops significantly affected binding affinity to TIA1. The authors then demonstrated that interfering with the interaction of SL3 and TIA1 by designing specific ASO substantially reduced the amount of SARS-CoV-2 RNA replication in Huh7.5.1 cells via experiments on virus-infected cells from laboratory.
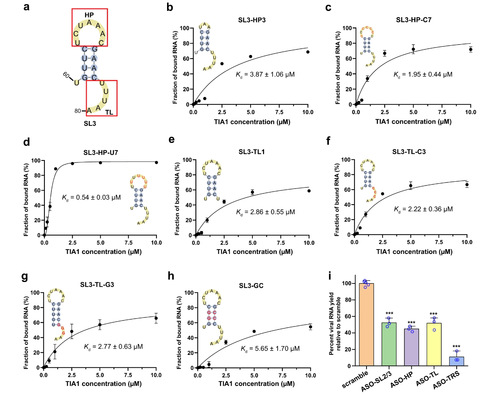
Figure 2. Binding capabilities of various SL3 RNA variants with human TIA1 protein. (a) WT SL3 consists of a 7-nt hairpin loop (HP), a 4-base pairs stem, and a 5-nt 3′-terminal loop (TL). The binding curves of human TIA1 protein with various mutated and truncated variants of SL3 RNA element: b SL3-HP3, the 7-nt HP reduced to 3-nt length, c SL3-HP-C7, sequence of 7-nt HP mutated to oligo C, (d) SL3-HP-U7, sequence of 7-nt HP mutated to oligo U, (e) SL3-TL1, last four nucleotides of 5-nt TL deleted, (f) SL3-TL-C3, three successive uridines in TL replaced by three cytosines, (g) SL3-TL-G3, three successive uridines in TL replaced by three guanines, h SL3-GC, two middle A-U base pairs in the stem substituted by G-C base pairs. All the secondary structures for SL3 variants are predicted by RNAStructure36 and mutated nucleotides are colored by red. All the Kd values were calculated from the EMSA image quantification from three independent experiments. Data are presented as mean±SD. (i) The yield of the bona fide SARS-CoV-2 virus with designed ASOs targeting SL3 RNA elements (including HP and TL loops and the transcriptional regulatory sequence (TRS)) in Huh7.5.1 cells for 24h, compared to the “scramble” control treated with a non-targeting ASO. SL2/3 has previously been reported18. Data represent the mean±SEM; n=3 biological replicates. ***p=3.4×10−6 (ASO-SL2/3), 4.4×10−7(ASO-HP), 1.2×10−5(ASO-TL), and 1.6×10−7(ASO-TRS) using unpaired two-sample Student’s t test.
Finally, given the highly conserved sequence of the cis-acting element in the 5'-UTR in the beta coronavirus RNA genome, the authors found that the human TIA1 protein also interacted with the SL3 element in coronaviruses such as MERS and SARS with comparable binding affinity.

Figure 3. Sequence comparison of SL3 RNA elements in betacoronavirus genomes and their binding abilities to human TIA1 protein. (a) Multiple sequence alignment showing nucleotide coverage within SL3 RNA elements of representative species within genus Betacoronavirus. The transcriptional regulatory sequences (TRS) are highlighted by red box. Six identified sites important for SARS-CoV-2 SL3 binding by TIA1 protein are marked by red triangles. Sequence positions of SL3 RNA element in each species are given in parentheses. BatHp, Bat Hp-betacoronavirus/Zhejiang2013; RousettusBat, Rousettus bat coronavirus; RousettusBatHKU9, Rousettus bat coronavirus HKU9; PipistrellusBat, Pipistrellus bat coronavirus HKU5; TylonycterisBat, Tylonycteris bat coronavirus HKU4; England1, Betacoronavirus England 1; MERS, Middle East respiratory syndrome-related coronavirus; Erinaceus, Betacoronavirus Erinaceus/VMC/DEU/2012. The binding curves between human TIA1 protein and SL3 RNA elements from (b) RousettusBatHKU9 and (c) MERS genomes. The Kd values were calculated from the EMSA image quantification from three independent experiments. Data are presented as mean±SD.
The work is supported by funds from the National Key R&D Program of China, the National Natural Science Foundation of China, National Independent Innovation Demonstration Zone Shanghai Zhangjiang Major Projects, the National Center of Technology Innovation for Biopharmaceuticals, Shanghai Artificial Intelligence Lab, the Starry Night Science Fund at Shanghai Institute for Advanced Study of Zhejiang University etc. To access the article please visit https://www.nature.com/articles/s41467-023-39410-8.

