Shanghai Institute for Advanced Study, Zhejiang University (SIAS, ZJU), together with Multiscale Research Institute for Complex Systems, Fudan University (MRICS, FU) and Centre of Computational Biology, Shanghai Artificial Intelligence Laboratory (CCB, PJ Lab), hosted 2023 Word Artificial Intelligence Conference (WAIC) AI for Science: Computational Biology Forum on 7 Jul 2023 at Shanghai World Expo Center.
Providing theoretical basis for biomedical research and development, computational biology marked one of the most prominent applications for AI technology. The emergence of AlphaFold2 and ChatGPT proves that AI-based algorithms have already surpassed traditional computational methodologies in certain aspect, and had found itself in various aspects in biomedical research, including high-throughput screening, targeted discovery and molecule prediction for drugs.
In the light of this, SIAS, ZJU joint efforts with MRICS, FU and CCB, PJ Lab, hosted the forum AI for Science: Computational Biology at 2023 WAIC in Shanghai, China. Keynote speakers for the forum include Michael Levitt, Nobel Laureate and professor at Stanford University, John Zhang, Professor from Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Feng Gai, Professor from Peking University, Haiyan Liu, Professor from University of Science and Technology of China, Brandon Xia, Professor from McGill University, and Jianpeng Ma, Professor from Fudan University. The roundtable panelists included Jin Wang from Stony Brook University, Yaoqi Zhou, Professor from Shenzhen Bay Laboratory, Yafeng Deng, CEO of CarbonSilicon Intelligence, and Xiang Zhao, Director of Algorithms fromTuring Quantum. The roundtable discussion was chaired by Ruhong Zhou, Professor from Zhejiang University and Director of SIAS, ZJU. Professor Kun Luo, Deputy Director of SIAS, ZJU and participants from leading pharmaceuticals and AI technology companies attended the forum.

Professor Jin Wang opened the forum by indicating that computational biology marked the evolution of biology from descriptive research to quantitative analysis, integrating multiple disciplines such as computer science, biology, big data, applied mathematics, chemistry and genetics. Further development of computational biology calls for further communications and collaborations between academics and industry.

Professor Michael Levitt delivered the opening talk, and he explained the impact of Biological Intelligence, Human Intelligence and Machine Intelligence on science, technology and innovation. Professor Levitt reviewed the history of study in protein folding modeling to reveal protein structure and function, and he illustrated the biological basis of AI and how neural networks such as ChatGPT can be constructed and used for learning. Professor Levitt ended his talk by indicating the trinity of connections between biology, humans and computers.

Professor Jianpeng Ma presented Computational Structural Biology: Algorithms to Applications. He introduced in detail how technological platform could support the process from new target proteins discovery, in silicon structure prediction, experimental structure determination, to simulation development and drug design. He believed that conformational flexibility is one of the most difficult barriers in improving resolution, and extracting dynamic information directly from experimental data is challenging. Professor Ma emphasized that by applying intelligent algorithms to resolve experimental data with poor signal-to-noise ratios, it was possible to directly extract dynamic information and construct 3D protein structures, which greatly improved the efficiency and precision of protein structure resolution. He pointed out that the algorithm seemed vital when facing restrictions from computer hardware.

Professor John Zhang shared his latest research on mutations in pMHC-TCR complex. Professor Zhang thoroughly introduced various free energy calculation methods, explaining their importance in studying protein-protein interactions and in understanding immune response, and how those methods provide new pathways to solve complex biological problems. He further demonstrated how to apply quantum chemistry (QM) to improve the accuracy in predicting protein interaction energies, and also explained ways to address scaling problem in QM.
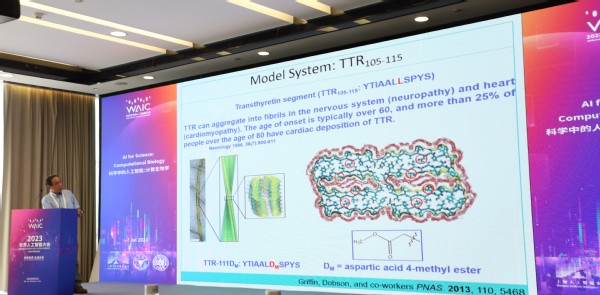
Professor Feng Gai talked about Spectroscopic Study of the Structure and Dynamics of Amyloid Fibrils. He discussed the combination of ultrafast vibrational spectroscopy and site-specific vibrational probes, and believed such strategy was key to understand biological problems, especially in the study of amyloid fibril structure and dynamics. Professor Gai further explored how AI could empower spectroscopic research from an experimental point of view to improve efficiency and accuracy. His insights inspired researchers to employ AI in biological experiments.

Professor Haiyan Liu explained Protein Design with Deep Learning, bringing a wide range of innovative data-driven approaches to design novel protein structures and sequences. He indicated that designing the backbone first and select sequence afterwards could be an effective way to do ab initio protein design. Professor Liu illustrated in detail how deep learning could compensate traditional design methodologies based on energy function, helping researchers to predict and design more accurately in data-intensive situations. He further shown that the models they trained have been experimentally validated in sequence selection and backbone generation, demonstrating great potential of deep learning in biomedicine.
Professor Ruhong Zhou chaired the second half of the Forum.

Professor Xia Yu presented on multi-scale modelling of biological networks. Systems biology builds a panorama of biological systems by describing the interactions and networks among proteins and other biomolecules within a cell. Professor Xia elaborated on the correlation between the rate of protein evolution and the physical properties of the sites where they are located. He further explained that proteins differ significantly in structure and function internally, externally and at interfaces, and were subjected to evolution pressure. By modelling protein interactions and comparing human protein-protein interfaces to that of viral proteins, Professor Xia found that viral proteins could mimic human proteins by precisely selecting their sites and evolving at a higher rate. These findings provided new insights into how viruses use human proteins to replicate and reproduce, and how to design new antiviral strategies. Professor Xia's talk enhanced the understanding of multi-scale biological network modelling, and demonstrated the value of such research in biological sciences.

A roundtable discussion was initiated after the presentations. The panelists included Professor Jin Wang from Stony Brook University, Professor Yaoqi Zhou from Shenzhen Bay Laboratory, Deng Yafeng from CarbonSilicon Intelligence corperation and Dr Xiang Zhao from Turing Q, and Professor Ruhong Zhou served as facilitator. Professor Wang and Professor Yaoqi Zhou elaborated on structure prediction for drug discovery from academics point of view, and explained how artificial intelligence and computational technology development facilitated drug screening. Mr Deng and Dr Zhao shared their experiences in academia-industry partnerships, especially in algorithms R&D and quantum computing. Professor Ruhong Zhou also invited keynote speakers and audiences to join the discussion. Perspectives from multiple educational and research backgrounds were shared and merged, connecting academia and industry.

