The ability of neurites of the same neuron to avoid each other (self-avoidance) is a conserved feature in both invertebrates and vertebrates. The key to self-avoidance is the generation of a unique subset of cell-surface proteins in individual neurons engaging in isoform-specific homophilic interactions that drive neurite repulsion rather than adhesion. Among these cell-surface proteins are fly Dscam1 and vertebrate clustered protocadherins (cPcdhs), as well as the recently characterized shortened Dscam (sDscam) in the Chelicerata.
Recently, Dr Yongfeng Jin, professor of College of Life Sciences, Zhejiang University, Adjunct Professor, Shanghai Institute for Advanced Study, Zhejiang University, and his collaborators published the review Confluence and Convergence of Dscam and Pcdh Cell-recognition Codes in Trends in Biochemical Sciences, reviewing recent advances in understanding of how cPcdh, Dscam, and sDscam cell-surface recognition codes were expressed and translated into cellular functions essential for neural wiring.
Fly Dscam1, vertebrate cPcdhs, and Chelicerata sDscams
Dscam1 and vertebrate clustered procalcitonin Pcdh are examples of single genes generating extreme isoform diversity, playing important roles in neuronal recognition assembly. In flies, Dscam1 potentially generates 19 008 distinct Ig-superfamily protein isoforms by mutually exclusive splicing. In humans, tandemly arranged clusters of three protocadherin gene clusters can generate 53 protocadherin gene clusters. Interestingly, in Chelonians, a hybrid gene family, sDscam, has recently been identified as sequence homologous to Drosophila Dscam1 but strikingly similar in genome assembly to vertebrate Pcdh. Dscam1 and Pcdh are able to mediate neuronal self-recognition and self-avoidance, providing a remarkable example of convergent evolution (Figure 1).
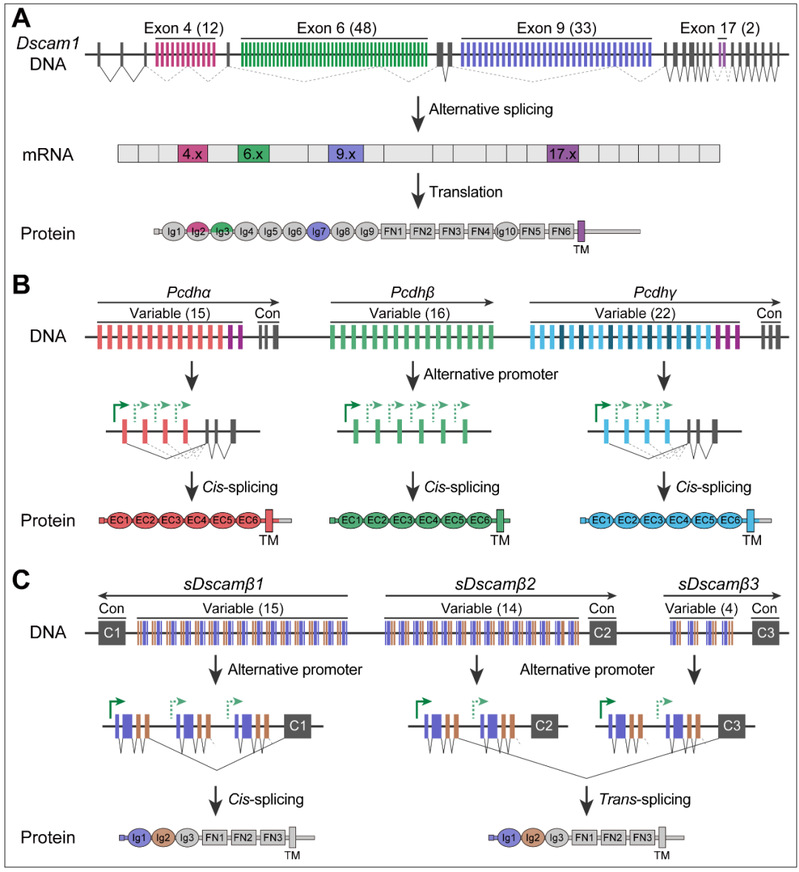
Figure 1: Genomic organization of fly Dscam1, human cPcdhs, and mite sDscams
Evolutionary origins of Drosophila Dscam1, vertebrate Pcdh, and chelonian sDscam
The Pcdh and Dscam genes originated prior to the divergence of symmetrical animals and cnidarians 600 million years ago, but underwent an evolutionary branch-specific expansion of subtype diversification. First, the pan-crustacean Dscam1 gene generated subtype diversity through mutually exclusive splicing of four variable exon clusters. Second, polypod species utilized gene duplication and mutually exclusive splicing to generate Dscam isoform diversity. Finally, chelonians have evolved a large number of truncated sDscams. sDscamα and sDscamβ genes have a series of tandem cassette repeats at the 5' end of the genes encoding one and two Ig structural domains, respectively, with variable cassettes ranging from 50 to 100. In vertebrates, mammals have species-specific expansion and contraction of the Pcdh gene. In contrast, Pcdh is not present in the genomes of molting animals, which have evolved complex Dscam, suggesting convergent evolution between arthropods and vertebrates (Figure 2).
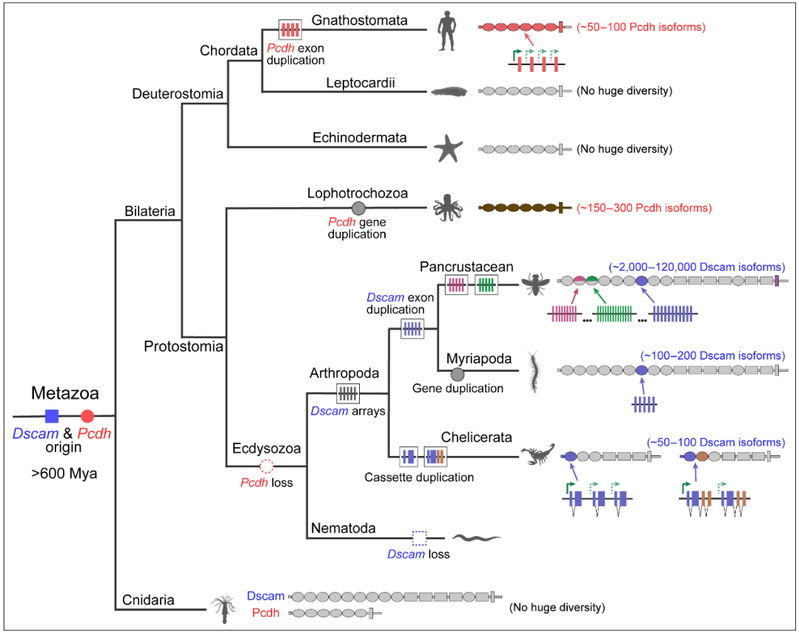
Figure 2: Distal cyclization regulate variable isoform expression of Pcdh, Dscam1, and sDscam
Distal cyclization mediates variable isoform expression of Pcdh, Dscam1, and sDscam
The three-dimensional genomic structure of Pcdh plays an important role in regulating gene expression. The mammalian Pcdh gene cluster consists of two sub-topology-associated structural domains (TADs), Pcdhα and Pcdhβγ. The sub-TADs of Pcdh are generated by CTCF-mediated chromatin interactions between distal enhancers and target promoters. Specifically, chromatin loops are formed by ATP-driven Cohesin protein-mediated ring extrusion and are preferentially generated between forward-reverse polymerized CTCF binding site (CBS) elements. All Pcdh gene clusters have tandem arrays of directed CBS elements in remote enhancers and their target promoters.
Competitive RNA pairing between anchor sites and selection sequences provided a mechanistic model for regulating mutually exclusive variable splicing of Dscam1. Disruption and compensatory mutations indicated that mutually exclusive splicing of Dscam1 variable exons 4, 6, and 9 was regulated by anchor-localization site-selection sequence base pairing. Competitive RNA pairing between anchor localization sites and selection sequences was recently identified for the 5' variable cassette exon of sDscamβ. Disruption and compensatory mutations indicated that splicing of individual variable cassette exons was regulated by anchor-localizer-selective sequence base pairing, and, RNA pairing between anchor-localizer and selective sequence molecules regulated trans splicing (Figure 3).
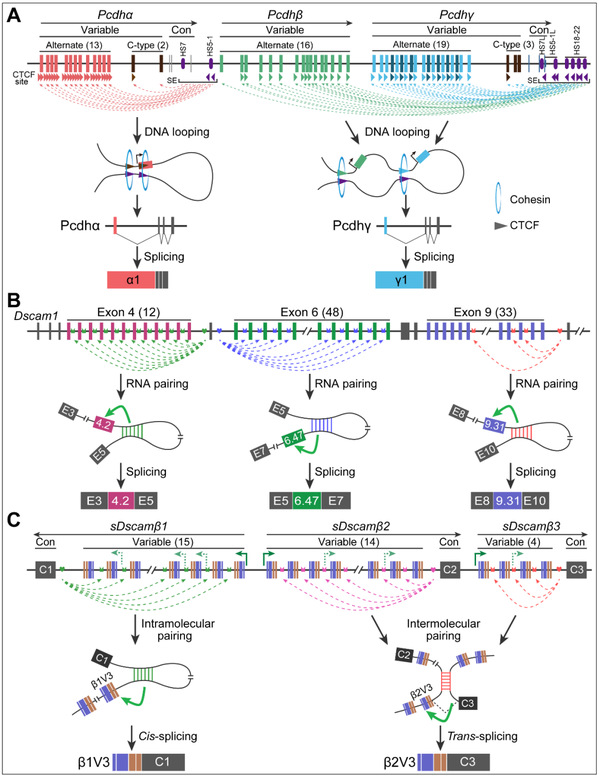
Figure 3: Long-distance looping mediates isoform expression in mammalian cPcdh, fly Dscam1, and chelicerate sDscam
Dscam1, Pcdh, and sDscam exhibit homology-specific trans-interactions
Drosophila Dscam1, vertebrate Pcdh, and chelicerate sDscam isoforms showed specific homologous trans-binding. In Drosophila Dscam1, such homologous binding was specifically dependent on the variable structural domains Ig2, Ig3 and Ig7. The Ig structural domains interacted in a modular fashion, and mismatches prevent binding. Similarly, trans-interactions between cis-dimers of the Pcdh heterodimers were strictly homodimeric and mediated by the extracellular domains EC1-EC4. For the cheliped sDscam, homologous trans-interactions were mediated by variable Ig1 domain, whereas variable Ig2 domain did not affect homologous binding (Figure 4).
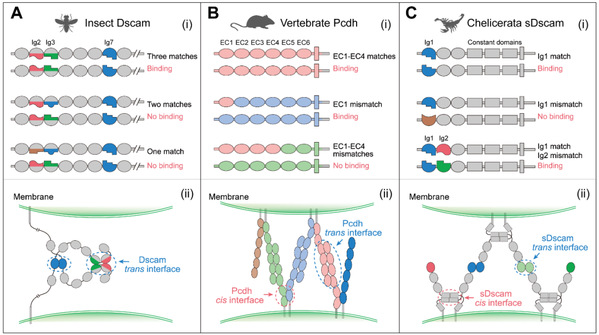
Figure 4: Fly Dscams, mammalian Pcdhs, and chelicerate sDscams exhibit isoform-specific homophilic binding
Dscam1 and Pcdh mediate dendritic self-avoidance and self/nonself discrimination
In Drosophila, Dscam1 produces tens of thousands of cell-surface proteins with isoform-specific binding. Each neuron stochastically produces different subsets of Dscam1 isoforms that can engage in highly specific homophilic binding between apposing cell surfaces, thereby conferring a cell identity code for self-recognition and self-avoidance. Deletion of the gene resulted in significantly reduced dendrite repulsion between self-branches. Reducing Dscam1 diversity lead to reduced dendritic overlap between class I and class III dendritic branching neurons. Thus, while expression of a single Dscam1 isoform was sufficient to maintain self-avoidance, thousands of isoforms were required to discriminate between self and non-self. Similarly, conditional knockdown of vertebrate Pcdhs led to dendritic collapse in astrocytes, a classic self-avoidance defect. Deletion of three Pcdh clusters resulted in severe axonal branching defects and loss of self-avoidance in mouse olfactory sensory neurons. Furthermore, deletion of the Pcdhα/γ cluster led to more severe survival and self-avoidance defects than deletion of Pcdhγ only, demonstrating Pcdh’s role of cell-surface identifier that mediates self-avoidance and self- and non-self discrimination in vertebrates. Recent studies in cortical neurons have reported a potential intracellular signaling pathway in which Pcdh mediates homologous binding (Figure 5).
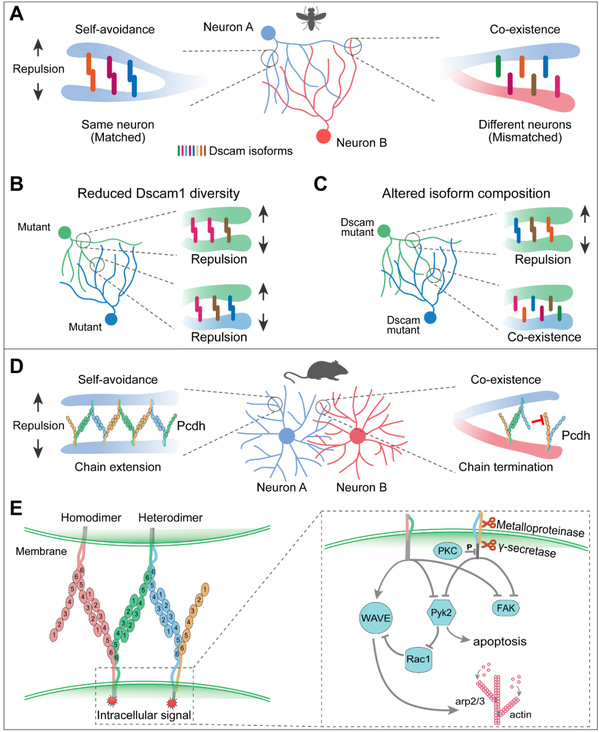
Figure 5: Dscam1 and cPcdh mediate dendritic self-avoidance and self/nonself discrimination
Beyond self-avoidance: challenging the classical view
Multiple genetic evidence confirmed Dscam1 mediated self-avoidance in dendritic branching neurons, but self-avoidance was a functional mechanism of Dscam1 in axon bifurcation in the mushroom body (MB) remains controversial. In traditional models, sister-branch axon segregation in MB neurons relied on Dscam1-mediated rejection signaling. However, recent genetic evidence has uncovered Dscam1 mutants exhibiting defective phenotypes: truncated MB lobes and thinning of both lobes, which could not be explained by Dscam1-mediated self-rejection. In addition, recent Dscam1 phenotype-diversity correlation analyses revealed that Dscam1 diversity was differentially required for the developmental patterns of dendritic branching neurons, mushroom bodies, and mechanosensory neurons. These results suggested that Dscam1 playled distinct roles in axons and dendrites via different mechanisms. In addition, members of the Pcdhα cluster also regulated cortical neuron migration, and its deletion resulted in spinal cord interneuron death and neonatal mouse lethality. Thus, Drosophila Dscam and mammalian Pcdh also exhibited non-self-avoidance functions in neuronal development (Figure 6).
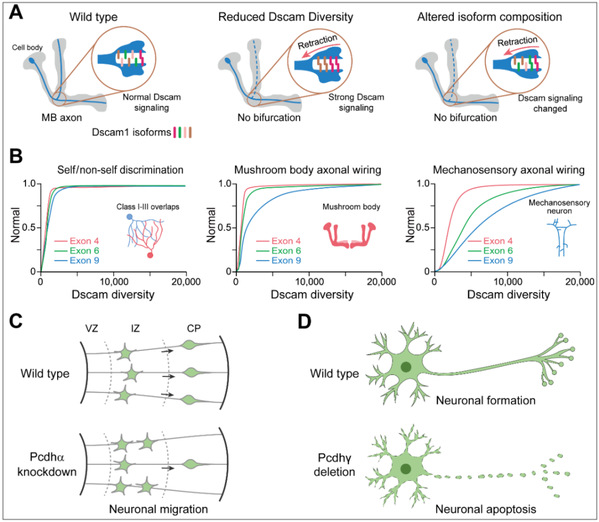
Figure 6: Fly Dscams and mammalian Pcdhs function beyond self-avoidance
In summary, extraordinary isoform diversity of fly Dscam1, sDscams, and cPcdhs as a unique code of cell-surface recognition proteins and their underlying molecular mechanism provided insight into gene expression regulation based on alternative promoters and alternative splicing. Recent structural and biochemical studies have revealed a zipper-like cell recognition model in chelicerate sDscams, suggesting that Chelicerata sDscams have more structural parallels with vertebrate cPcdhs than with fly Dscam1, and exploration of the noncanonical functions of Dscam1 and cPcdh diversity in neuronal wiring has also enhanced our understanding of their functions beyond self-avoidance.
The work could be accessible at https://www.cell.com/trends/biochemical-sciences/fulltext/S0968-0004(23)00227-X.

