The complexity and orderliness of the brain has always been a hot but difficult topic for life science researchers. Neurons perceive external world and control biological behaviours by establishing precise connections to form specific neural circuits and synapses, which requires the neurons to be able to recognise self and non-self. Neurons are endowed with unique cell surface identities through the expression of diverse recognition molecules, such as Down syndrome cell adhesion molecule (Dscam) and mammalian clusters Protocadherin (cPcdh) to realize specific recognition.
There are 38,016 isoforms of Drosophila Dscam and 52 isoforms of human cPcdh, and it has been puzzling that the two genes share similar functions in spite of the huge difference in the number of isoform. Recent work found that there existed a special shortened Dscam (sDscam) in chelonians. sDscam has a protein structure similar to that of Drosophila Dscam, and its gene is organized in a more similar to that of cPcdh in vertebrate. The number of isoforms of sDscam in chelonians is around 100, and it displayed the features of both Dscam and cPcdh in terms of isomer generation (Fig. 1). Therefore, sDscam is considered to be a transition gene between Dscam and cPcdh, providing a possibility to explain the evolutionary gap between Dscam and cPcdh.
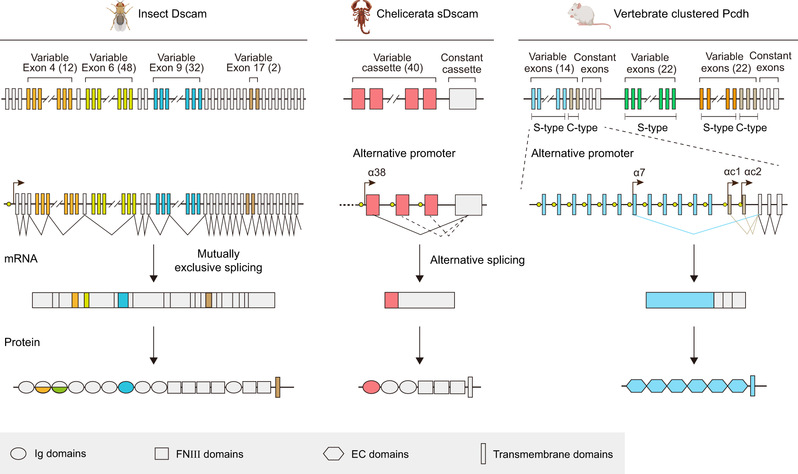
Fig. 1: Drosophila melanogaster Dscam, Mesobuthus martensii sDscam and mouse cPcdh have been chosen as samples. The variable regions are indicated by different colors, while the common regions are colored in gray. The promoters are represented as yellow filled circles. Dscam applies mutually exclusive RNA splicing to generate tens of thousands of isoforms. For cPcdh, each variable exon is preceded by a promoter and only the cap-proximal exon engages in the splicing with the first constant exon. The shorten sDscam uses both alternative promoter and RNA splicing to generate its isoform diversity. For clarity, only the α subfamilies are shown for sDscam and cPcdh. More details of sDscam isoforms are shown in Supplementary Fig. 1. The species icons are created with MedPeer (www.medpeer.cn).
Recently, Dr Yongfeng Jin, professor of College of Life Sciences, Zhejiang University, Adjunct Professor, Shanghai Institute for Advanced Study, Zhejiang University, and his collaborators published an article Structural Basis for the Self-recognition of sDSCAM in Chelicerata in Nature Communications, revealing the molecular details of the neuronal self-recognition of neuronal surface receptor sDscam through trans and cis interactions, and suggested that sDscam may be the missing link in the evolutionary transition from insect Dscam to higher mammalian cPcdh.
The authors analysed the crystal structures of 12 sDscam protein fragments and revealed two types of sDscam interactions: trans interactions and cis interactions. The trans interactions between different cells were mediated by homodimers formed by the Ig structural domain of the sDscam molecule, which were heterodimer-specific and acted in a manner similar to that of Drosophila Dscam. The cis interactions on the surface of the same cell were mediated by the FNIII structural domain, which was similar to that of mammalian cPcdh. On the cell surface, sDscam was arranged in a zipper-like model (Fig. 2) in a way similar to mammalian cPcdh, so that a limited number of isoforms could be used to differentiate between a large number of cells.
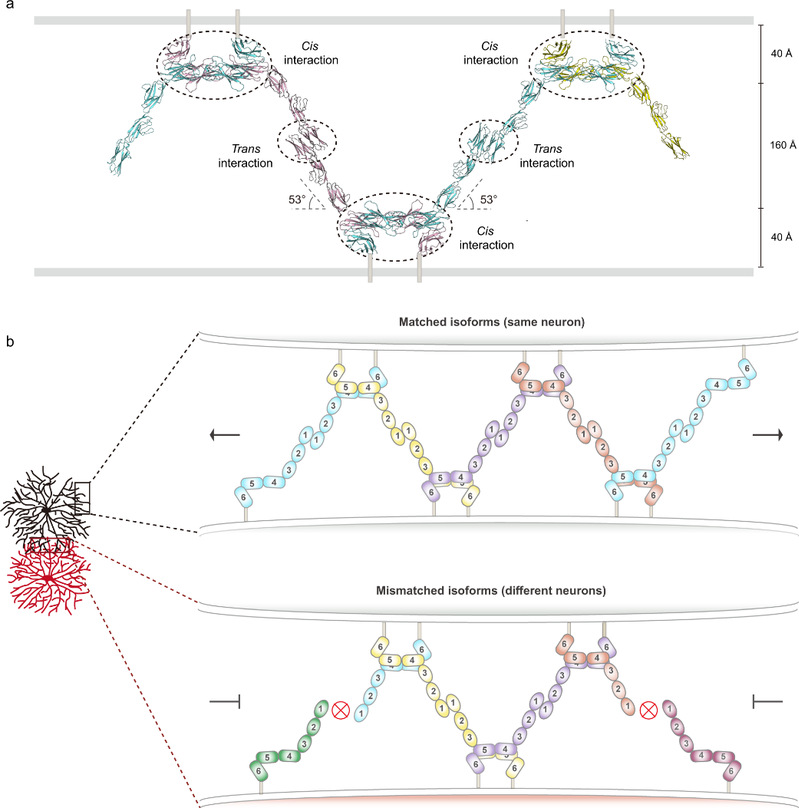
Fig. 2: a) Schematic of the zipper-like structure of sDscam molecules between apposed membrane surfaces. The domains involved in the cis or trans interactions are indicated by dashed circles. b) Proposed zipper-like model for sDscam recognition. Chain extension of sDscam molecules occurs between the dendrites of the same neuron which have the same isoform repertoire, resulting in the sDscam-mediated cell-cell recognition. A switch-like repulsion response might be elicited when the extension of the zipper reaches a threshold. On the contrary, dendrites of different neurons owning mismatching isoforms will terminate chain extension.
On cell surface, two neighboring sDscam molecules formed a cross cis-dimer via the FNIII domains in a side-by-side manner, while each extended arm (Ig domains) of a sDscam cis-dimer engaged in trans with two different cis-dimers on an apposed cell in a hand-in-hand manner. Thus, a continuous array assembled through alternating cis and trans interactions of sDscam. In order to incorporate into the zipper-like assembly, the incoming sDscam cis-dimer must have one isoform to match the isoform on the exposed end of the zipper assembly. The zipper assembly wiould terminate when a matched isoform is not available, resembling the chain-termination model for cPcdh-mediated discrimination between self and non-self.
The above structural and functional results reported showed that sDscam Ig1 mediated trans interaction resembling insect Dscam1 Ig7, while the membrane-proximal FNIII domain mediated cis interaction and might form a zipper-like assembly mimicking the vertebrate clustered Pcdh. Together, these studies advanced our understanding of sDscam and shed light on the evolutionary landscape of the recognition molecule diversity. The work could be accessible at https://www.nature.com/articles/s41467-023-38205-1.

