Bacteria have evolved various response systems to adapt to environmental stress. Most members of Deinococcus display superior DNA damage repair capabilities and are critical to study on genome stability maintenance mechanisms. Previous studies showed a protease-based derepression mechanism in response to DNA damage was controlled by the specific cleavage of repressor DdrO by metallopeptidase PprI (also known as IrrE). Despite the efforts to document the biochemical, physiological, and downstream regulation of PprI-DdrO, the upstream regulatory signal activating this system remains unclear.
On 29 February 2024, Professor Ruhong Zhou from Shanghai Institute for Advanced Study, Zhejiang University, together with Professor Yuejing Hua and Professor Ye Zhao from College of Life Sciences, Zhejiang University, together published an article 'The Deinococcus protease PprI senses DNA damage by directly interacting with single-stranded DNA'. The work identified DNA damage sensors and revealed that single-stranded DNA (ssDNA), a product of DNA damage, could serve as the signal for DNA damage sensing in the metalloprotease/repressor system in bacteria by initiating transcription of relevant DNA damage repair genes.

Previous studies have shown that the transcript levels of PprI gene did not differ significantly before and after DNA damage, suggesting PprI protein did not activated by increased gene expression. Here the authors first found that ssDNA could bind to PprI protein and activated its cleavage. They also revealed structure of the PprI-ssDNA complex via combing biochemical, cellular and structural biology research. They further located three binding sites crucial to the activation of intracellular PprI-DdrO system (Figure a). Next, they studied the structure of the PprI-ssDNA-DdrO complex by computational biology methods, and concluded that ssDNA activated transcriptional levels of downstream genes by enhancing the interaction between PprI and DdrO (Figure b). In addition, the dynamic equilibrium between the monomer and dimer of the PprI protein was involved in the activation (Fig. c).
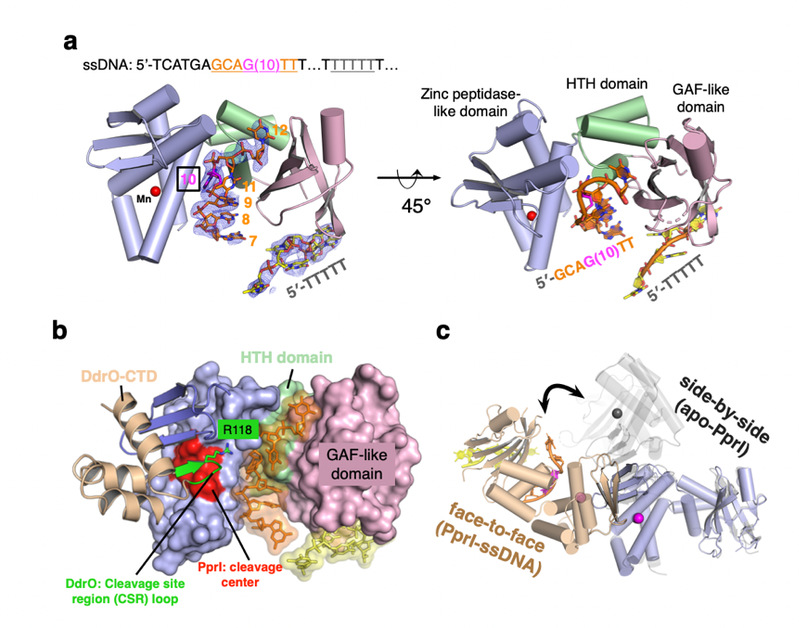
a) Overall structure of PprI-ssDNA complex. DG-PprI and ssDNA are labeled and shown as cartoon and sticks, respectively. A schematic of the DNA substrate used for crystallization is shown on top with colors corresponding to those observed in the PprI-ssDNA structure below. The electron density of two segments of ssDNA (5ʹ-GCAGTT and 3ʹ-TTTTT) is shown in blue with the refined 2Fo-Fc contoured at 1σ. The catalytic metal ion is shown as sphere and colored red. b) Snapshot of all-atom molecular dynamics (MD) simulations of the PprI-DdrO-ssDNA model. The DG-PprI, DdrO, and ssDNA are labeled and shown as surface, cartoon and sticks, respectively. The cleavage center of DG-PprI and the CSR loop of DdrO (Arg118) are highlighted in red and green, respectively. c) Superposition of two PprI dimer configurations with distinct colors (face-to-face dimer in slate and wheat; side-by-side dimer in black).
The work demonstrated that single-stranded DNA physically interacted with PprI protease, which enhanced the PprI-DdrO interactions as well as the DdrO cleavage in a length-dependent manner both in vivo and in vitro. Structures of PprI, in its apo and complexed forms with single-stranded DNA, revealed two DNA-binding interfaces shaping the cleavage site. Moreover, the dynamic monomer-dimer equilibrium of PprI was also important for its cleavage activity. These results also shed light on the survival and acquired drug resistance of certain bacteria under antimicrobial stress through a SOS-independent pathway, and could potentially facilitate the development of drugs tackling antimicrobial resistance.
To learn more please visit https://www.nature.com/articles/s41467-024-46208-9.

