Ubiquitination plays essential roles in eukaryotic cellular processes. The effector protein CteC from Chromobacterium violaceum blocks host ubiquitination by mono-ADP-ribosylation of ubiquitin (Ub) at residue T66. However, the structural basis for this modification is unknown. Recently, Dr Yongqun Zhu, Professor of Zhejiang University, Adjunct Professor of Shanghai Institute for Advanced Study, Zhejiang University, together with collaborators, published an article “Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs in Nature Chemical Biology. By resolving the structure of the pathogenic bacterial effector protein CteC complexed with ubiquitin in different catalytic states, and further biochemical and evolutionary biological analyses, the authors revealed the molecular mechanism by which CteC specifically recognizes ubiquitin and modifies it with ADP-ribosylation, and found that CteC represents a class of pathogenic bacteria that are widely present and a novel D-E family of ADP-ribosyltransferases ( ART).

ADP-ribosylation is a versatile post-translational modification present in all kingdoms of life, which is catalyzed by members of the ADP-ribosyltransferase (ART) superfamily of proteins. ADP-ribosylation is catalyzed by ADP-ribosyltransferases (ARTs), which transfer the ADP ribosyl group in the ligand molecule nicotinamide adenine dinucleotide (NAD+) to the substrate. Ribosyltransferases (ARTs) are catalyzed by ADP-ribosyltransferases (ARTs), which modulate the substrate function by transferring the ADP-ribose moiety from ligand molecule nicotinamide adenine dinucleotide (NAD+) to the substrate, thereby affecting the activity, stability, or interactions with other proteins of substrate protein.
Pathogenic bacteria inject effectors into host cells during infection via secretion systems. These effectors often target host cell signaling molecules, manipulate signaling pathways, and inhibit host immune defense responses to promote pathogen infection. Previous studies reported that the type III secretion system effector CteC from Chromobacterium violaceum specifically modified Ub via unusual threonine ADP-ribosylation on the residue T66, which impaired poly-Ub synthesis and caused dysfunction of poly-Ub chains in host cells. However, the catalytic mechanism of CteC and structural basis for this modification are unknown.
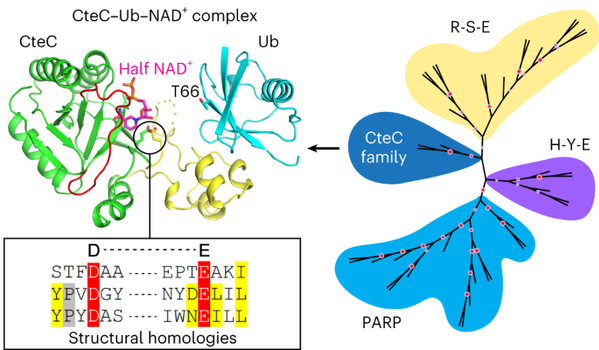
In this study, the authors determined the structures of CteC-Ub, CteC-Ub-NAD+ and CteC E220A-Ub-NAD+ complex, and found that the three-dimensional structure of CteC consists of relatively independent substrate-recognition and ART enzyme-activation domains. The substrate recognition domain binds to the I44 region of ubiquitin mainly through hydrophobic interactions. The enzyme-activated domain binds to the phosphate and nicotinamide groups of the ligand molecule, NAD+, in a nonclassical half-ligand binding mode. CteC undergoes a conformational change upon recognition of ubiquitin through the substrate recognition domain, thereby approaching the ubiquitin T66 and utilizes the catalytic amino acids D134 and E220 to mediate the cleavage of NAD+ and ultimately transfer the ADP-ribose moiety to ubiquitin.
In previous studies, ADP-ribosyltransferases have been divided into two major families, R-S-E and H-Y-E, based on the catalytic conserved amino acid motifs and structural folding patterns. In this study, after obtaining CteC structure, evolutionary biology analyses further revealed that the structural folding pattern was closer to that of the H-Y-E family, in particular the eukaryotic PARP family of ARTs in the H-Y-E family, compared with that of the R-S-E family of ARTs. Structural comparison with eukaryotic PARP2-HPF1 showed that D134 and E220 of CteC corresponded to E284 of HPF1 and E545 of PARP2, respectively, forming a unique ‘D-E’ motif. Through further structural homology searches, this study revealed that proteins structurally similar to CteC with conserved D-E catalytic motifs are widely found in a variety of microorganisms including bacteria and eukaryotic microorganisms, and demonstrated that such proteins all possess ADP-ribosyltransferase activity, suggesting that the D-E family of ARTs may play a more extensive biological function in microorganisms.
The structural homologs of CteC are found in more than 20 bacterial species, including commensal bacteria, suggesting that the ‘D-E’ family of ARTs are extensively involved in the pathogenesis or cellular processes of these bacteria. Interestingly, unicellular eukaryotes, including Haptolina ericina, also possess CteC-like ARTs. The wide distribution of the CteC-like ARTs in prokaryotic and eukaryotic microorganisms suggests that the ‘D-E’ family members may have the ability to modify residues other than threonine. In addition, the physiological role of CteC in blocking host ubiquitination during infection had been elucidated. Structural analyses with biochemical results reveal that CteC represents a large family of poly (ADP-ribose) polymerase (PARP)-like ADP-ribosyltransferases, which harbors chimeric features from the R-S-E and H-Y-E classes of ADP-ribosyltransferases. The family of CteC-like ADP-ribosyltransferases has a common ‘D-E’ catalytic consensus and exists extensively in bacteria and eukaryotic microorganisms.
This work was supported by grants from the Fundamental Research Funds for the Central Universities, NSFC and the National Science and Technology Major Project etc. To access the full article please visit https://www.nature.com/articles/s41589-023-01475-3.

